Originally posted in ELIXIR
Building on the success of the 2018 ELIXIR Implementation Study on BioContainers, an open-source registry for over 81,000 life science research related containerised tools, ELIXIR has launched three follow-up projects to progress and maintain the work in this valuable area. ❡
What are containers and why are they important?
Data produced by life science research is always on the increase. Likewise there has been a huge increase in the number of tools and platforms required to support data intensive analysis. These tools, code, back-end software and compute environments come in many flavours meaning it can be challenging to select the appropriate tools and run them smoothly in a local setting.
Software containers package up code and related dependencies so that software applications can run quickly and reliably from one computing environment to another. As an analogy, it’s like buying flat-pack furniture from IKEA: you have the instructions, components and tools all provided in a single package - enabling you to carry out the task (build the furniture) without having to source the required tools (screws, allen keys, instructions etc.).
This is an incredibly useful method of sharing analysis pipelines, software tools and documentation with collaborators and the wider scientific community thereby enabling reproducibility. Researchers can run the analysis without having to install dependencies and worry about the specific version of software that the person who originally created the pipeline used.
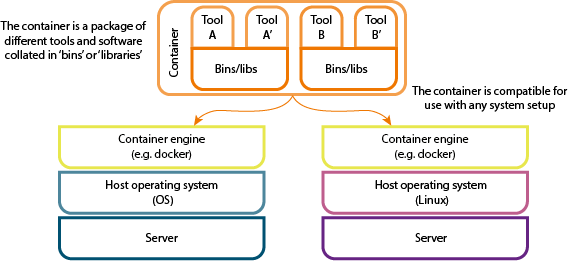
The BioContainers Registry
In 2018 ELIXIR launched a six month Implementation Study that aimed to build an infrastructure to help scientists within ELIXIR publish their software containers in a standardised manner. The result was the development of an open-source registry for life science research related to containers called BioContainers, which provided over 8,100 containerised tools. BioContainers is a community-driven project that provides the infrastructure and basic guidelines to create, manage and distribute bioinformatics packages (e.g. conda) and containers (e.g docker, singularity).
ELIXIR’s ongoing support for life science software containerisation
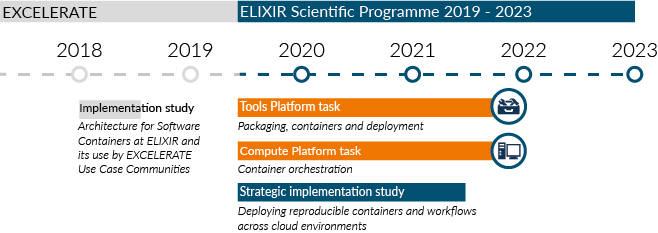
Three new tasks which will progress the work of the 2018 implementation study that developed BioContainers were detailed in ELIXIR’s Scientific Programme 2019 - 2023:
- The ELIXIR Tools Platform has a focused set of aims within the Packaging, containers and deployment task to maintain and extend the function of BioContainers.
- The ELIXIR Compute Platform has a dedicated set of tasks on Container Orchestration to enable standardised containers to be deployed across the Nodes.¶
- A Strategic Implementation Study linking national infrastructure in twelve Nodes with the ELIXIR Compute and Tools Platforms developments. The project, Deploying Reproducible Containers and Workflows Across Cloud Environments, is driven by use case applications in single cell transcriptomics, metabolomics, proteomics and access-controlled human genomics.
“We’re really pleased that ELIXIR has chosen to continue support for the BioContainers project,” says Tools Platform task lead Yasset Perez-Riverol of EMBL-EBI, “We’re looking forward to putting our ideas and plans for BioContainers into action. ELIXIR’s support means that we’ll be able to continue supporting those working in life science research to carry out complex analysis in a more streamlined way through the use of software containers.”
Bringing together experts from across Europe to work collaboratively to solve the challenges in computational biology is one of the core foundations on which ELIXIR was founded. In total, seventeen ELIXIR Nodes are working across these three projects. By offering funding and organisational support, ELIXIR is able to facilitate this research that would otherwise be a challenge to achieve. The sum total of investment contributed by ELIXIR to the Implementation Studies and Platform tasks focusing on the area of containers amounts to approximately €1.6 million.
The driving force behind these projects is to ensure that scientists across Europe can execute the same set of tools reproducibly in different localities and the national cloud environments linked to national Nodes. There are four broad aims that these projects will address:
- Maintenance of BioContainers
This aspect will focus on monitoring the current usage of containers, and building up a picture of who is using them so that the correct support can be put in place. There are a lot of containers already in BioContainers, and any new updates or functionality generated by work in these two projects will also be applied to these.
- New functionality for BioContainers
While the 2018 Implementation Study was a great success, there are always new technologies and use-cases that need to be accommodated for. The activities in the Tools Platform tasks will focus on incorporating new container formats and specifications, as well as working to build containers automatically with software such as conda-forge. The Strategic Implementation Study will take a more focused approach into how BioContainers can be integrated into cloud environments, particularly the EOSC-Life workflows which utilise Galaxy and NextFlow.
- Hybrid Cloud deployment of BioContainers
There is a strong drive for federated software containers, so that sensitive data or data that is restricted to certain countries can be handled and analysed securely. The Compute Platform’s task on Container Orchestration is developing a common method of deploying these containers on cloud computing resources across the various ELIXIR Nodes. This will be compatible with ELIXIR AAI identity and access management and, by co-developing GA4GH cloud standards, ultimately usable on related life science infrastructures worldwide.
- Community building
Of course, there’s nothing to be gained in developing anything in isolation from the communities that will use and help build this resource. The strategic implementation study’s plans include organisation of hackathons and training events, as well as developing containers and workflows for ELIXIR Communities with Galaxy and CWL.
Further information¶
The 2018 Biocontainers Implementation Study was led by Yasset Perez-Riverol, EMBL-EBI, and involved nine ELIXIR Nodes. The new cross platform Strategic Implementation Study is led by Steven Newhouse (EMBL-EBI) and Salva Capella Gutierrez (BSC, ELIXIR Spain) and brings together twelve Nodes. The project started on 1 June 2019 and will be completed by 31 May 2021.
You can find out more about the BioContainers software in a recent publication, and in this ELIXIR webinar. Should you like to find out more about the ELIXIR Tools Platform and its activities, please get in touch with the Tools Platform Coordinator Jen Harrow. The ELIXIR Compute Platform is coordinated by Jonathan Tedds.
Mon 11 November 2019
No comments:
Post a Comment